Model Lipid Bilayers
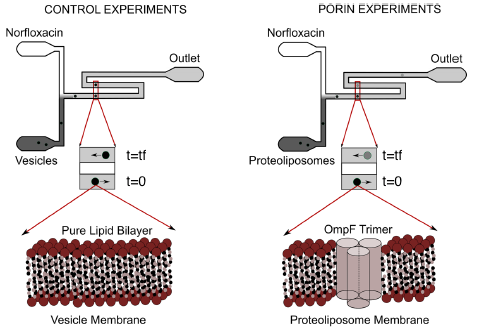
Cell membranes are the major interface between cells and their environment. In order to understand the physical mechanisms governing molecular flow we use model lipid bilayers such as giant unilamellar vesicles (GUVs) and lipid vesicles embedded with protein transporters. We control the physico-chemical environment around the vesicles by using bespoke microfluidic devices that also allow us to position individual vesicles and to create controlled gradient of substrates such as drugs or nutrients. We use epifluorescence microscopy to image substrate accumulation in single vesicles and image analysis and mathematical modelling to measure the parameters characterising transport kinetics, for example membrane permeability.
By using this approach and the autofluorescence of quinolone antibiotic drugs under ultraviolet illumination, we have developed a label-free microfluidic assay to quantitatively study antibiotic diffusion through lipid membranes (Cama et al. 2014, Lab on a Chip 14: 2303)
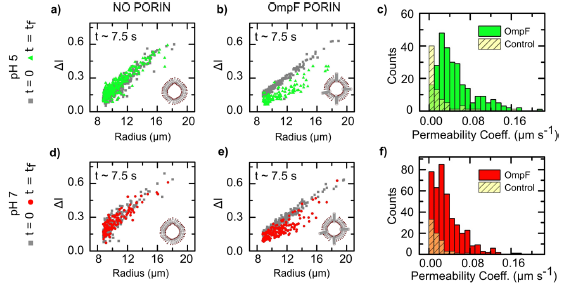
We showed that the environmental pH has a major impact on the permeability of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) lipid membranes to the quinolone norfloxacin, permeability values showing a 6-fold decrease in basic solutions (pH=5).
Furthermore, we studied the effect of the Escherichia coli outer membrane channel OmpF on the accumulation of norfloxacin in proteoliposomes. We showed that both at pH 5 and 7 At, the porin significantly enhance drug permeation across the proteoliposome membranes. Specifically, at pH 5, where norfloxacin permeability across pure phospholipid membranes is low, the porins increase drug permeability by 50-fold on average (Cama et al. 2015, Journal of the American Chemical Society 137: 13836).
We then expanded our measurements on a range of fluoroquinolone antibiotics and
found that their pH dependent lipid permeability can span over two orders of magnitude (Cama et al. 2016, Scientific Reports 6: 32824).
We are currently establishing a novel approach to investigate the transport of nutrients across lipid membranes mimicking those found in the different branches of the tree of life.
News
- 8-September-2023: Congratulations Erin! New paper out at ISME Communications
- 10-April-2023: Congratulations Ula! New paper out at PLoS Biology
- 7-June-2022: Congratulations Ula! New paper out at eLife



