Membrane Transport
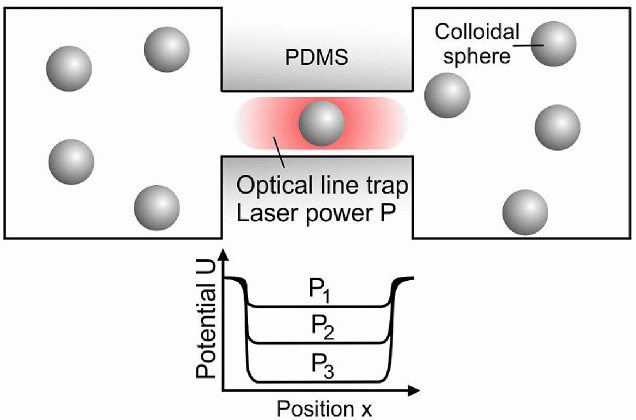
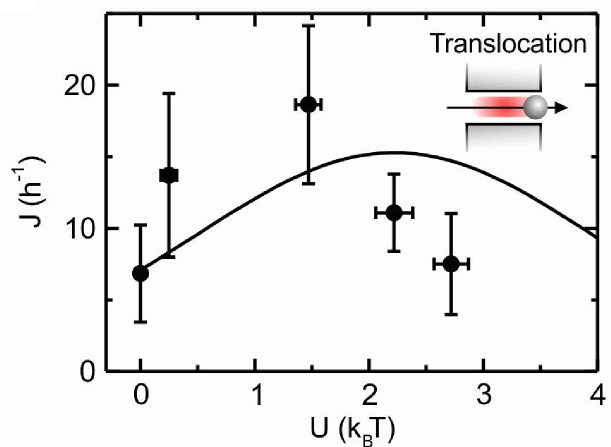
Transport of ions, proteins, antibiotics and other macromolecular solutes through channels and pores is ubiquitous in nature (Pagé et al. 2008, Nature Review Microbiology 6: 893) Channel-facilitated diffusion relies on optimized binding sites for the transported particles inside the channel. A few theoretical studies have rationalized channel-facilitated transport predicting a maximum in the diffusive current with respect to the binding potential (Berezhkovskii and Bezrukov 2005, Biophysical Journal 88: L17).
In order to validate these theories, we built a fully controllable platform combining microfluidics, microscopy and optical tweezers that allowed us to create a synthetic membrane channel that is an analogue of membrane protein pores through which molecules diffuse. By using this platform and in collaboration with Ulrich Keyser (Cavendish Laboratory, Cambridge) we have provided the first experimental evidence about the existence of an optimal strength in the binding between the membrane transporters and the transported particles (Pagliara et al. 2013, Advanced Materials 25: 844). We have also demonstrated that such binding maximizes particle transport, validating previously reported theoretical models about channel-facilitated diffusion. Furthermore, we have also discovered that the presence of binding sites at the transporter entrances, allows for an enhancement in particle transport up to 40 times higher with respect to free diffusion. This shed a light on the function of binding sites for transported molecules found at the mouths of a variety of transporters (Pagliara et al. 2014, Physical Review Letters 113: 048102). We have demonstrated that particle diffusion in confining channels is both hindered and anisotropic ( Dettmer et al. 2014, Physical Review E 89: 062305), which is relevant for quantitative models of channel-facilitated membrane transport. Moreover, in collaboration with Eric Lauga (DAMTP, Cambridge) we found that particle-particle interactions extend over the full channel length and have a constant strength that does not decay with particle separation. We explain the coupling mechanism using hydrodynamics with both a qualitative analytical model and quantitative comparisons with the
numerical simulations (Misiunas et al. 2015, Physical Review Letters 115: 038301).
In collaboration with the group of Rod Lim (Biozentrum, Basel), we have successfully reconstituted the two dimensional diffusion of colloidal particles on a molecular brush surface composed of nucleoporins - intrinsically disordered proteins that facilitate selective transport through nuclear pore complexes in eukaryotic cells - ( Schleicher et al. 2014, Nature Nanotechnology 9: 525 ). In collaboration with Matteo Pierno (Universitàs di Padova), we have demonstrated that bias forces as tiny as 1 fN determine the magnitude of particle escape time by drastically reducing interparticle collisions and switching off excluded-volume effects (Locatelli et al. 2016, Physical Review Letters 117: 038001).
We then translated this approach by using Escherichia coli as a model system for understanding the environmental influence on the capabilities of a single microbe to take up molecules.
The double-membrane cell envelope of Gram-negative bacteria is a formidable barrier to intracellular antibiotic
accumulation. A quantitative understanding of antibiotic transport in these cells is crucial for drug development,
but this has proved elusive due to a dearth of suitable investigative techniques.
Combining
microfluidics and time lapse autofluorescence microscopy we can rapidly quantify antibiotic
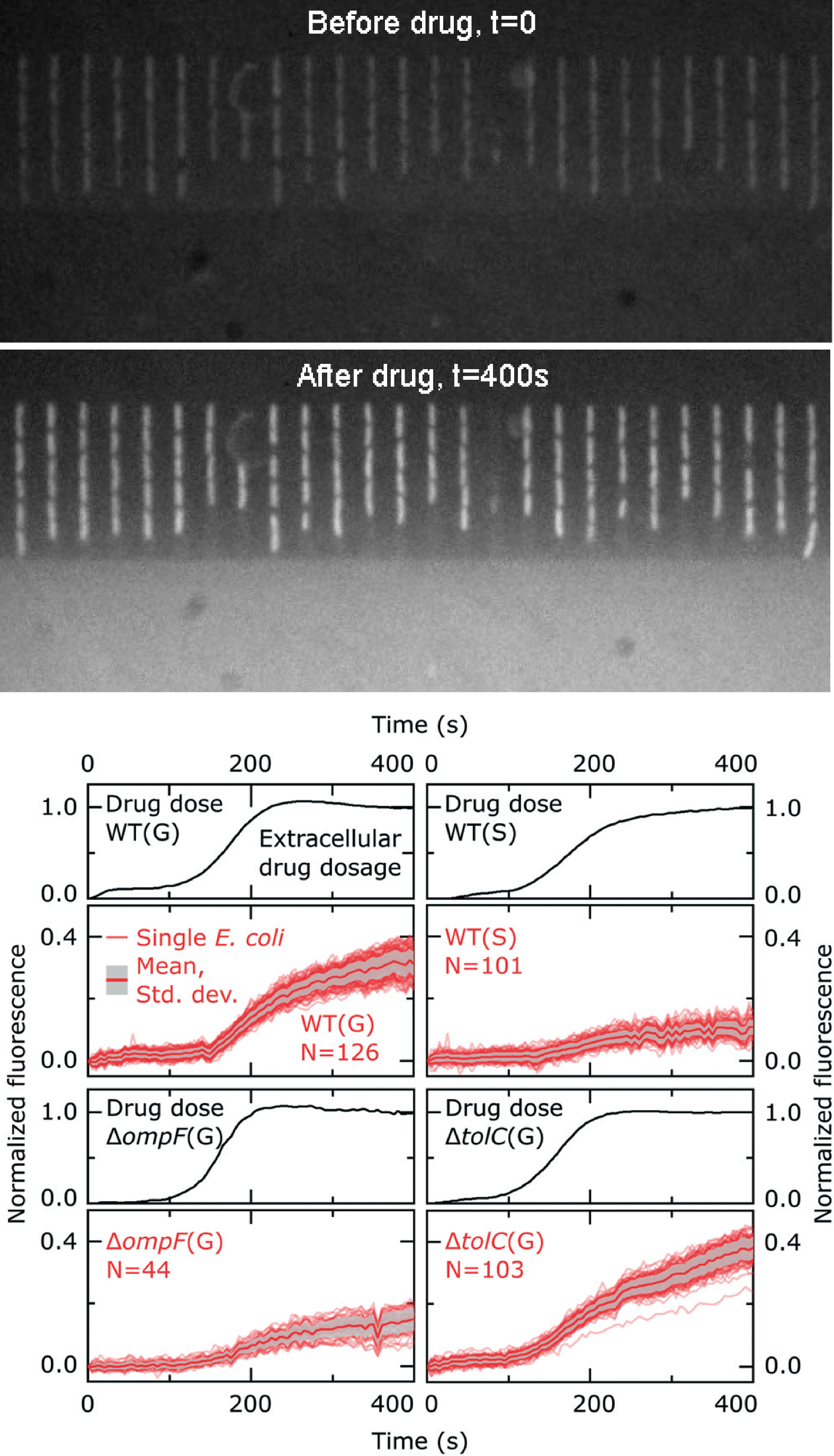
accumulation in hundreds of individual Escherichia coli cells. In fact, upon adding to the microfluidic environment autofluorescent antibiotics, such as ofloxacin, bacteria increase their internal fluorescence due to the progressive intracellular accumulation of antibiotics (see fluorescence images before and after ofloxacin addition on the left).
By serially manipulating the microfluidic environment,
we demonstrated that stationary phase Escherichia coli (wild type, WT, stationary, S, in the graphs on the left), traditionally more refractory to antibiotics
than growing cells (wild type, WT, growing, G, in the graphs on the left), display reduced accumulation of the antibiotic ofloxacin compared to actively
growing cells.
Our novel microfluidic method facilitates the quantitative comparison of the role of the
microenvironment versus that of the absence of key membrane transport pathways in cellular drug accumulation (e.g. the outer membrane porin ompF and the efflux pump tolC in the graphs on the left). Importantly, we performed for the first time a direct, quantitative
comparison of the role of growth phase versus the
role of the presence of individual transport pathways in
drug accumulation. We found that that the
growth phase plays an even bigger role in ofloxacin accumulation
than the complete removal of a major pathway for
drug accumulation that has often been linked to antibiotic
resistance
Unlike traditional techniques, our assay is rapid, studying accumulation as the cells are dosed with
the drug. This platform provides a powerful new tool for studying antibiotic accumulation in bacteria,
which will be critical for the rational development of the next generation of antibiotics. (Cama et al. 2020, Lab on a Chip 20: 2765).
