NEWS
Where it all happened - the locations where people have worked with lampbrush chromosomes worldwide since 1882
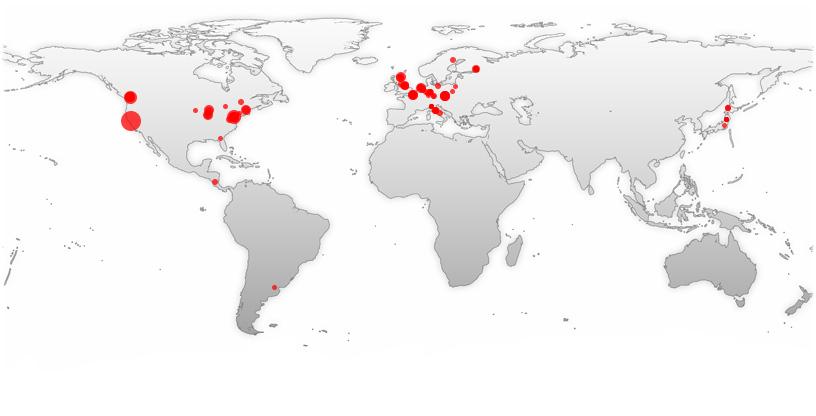
Where it all happened - the locations where people have worked with lampbrush chromosomes worldwide since 1882

(1) The Krasikova lab in St. Petersburg now has a new website (http://cell-nucleus-lab.bio.spbu.ru/).
(2) There are two interesting new protocol papers, one from the Gaginskaya Lab in St. Petersburg (Saifitdinova et al. 2017) on the handling of LBCs from birds and reptiles - exceedingly skilful but don't let this deter you from having a go!
The other from Garry Morgan (Nottingham, UK), published in Cold Spring Harbor Protocols 2018(4), full details have been added to our Protocols page (doi:10.1101/pdb.prot096958).
(3) Two new news-worthy publications. Elena Gaginskaya's team in St. Petersburg have now succeeded in characterising the repeat sequences that make up each of the 6 major chromomeres on the chicken W lampbrush chromosome - a data set that is especially significant in relation to the nature of the lampbrush chromomere (Komissarov et al. 2018).
Garry Morgan of Nottingham, UK, has published a really beautiful piece of work that adds some interesting and challenging information on the dynamics of lampbrush loop transcription and transcript processing. Read and think carefully about it (Morgan 2018)!
One year on the debate remains open, inviting your ideas and views. What are lampbrush chromosomes actually doing? Are they intensively transcribing from thousands of genes for some highly sophisticated reason that is connected to the requirements of the embryo or are they doing something else that perhaps has nothing to do with the fruits of their transcriptive endeavours? The oldest question in the field and still unresolved. Current RNA sequencing work in the Gall Lab in Baltimore may provide the answer but just suppose it doesn't. We're waiting and watching.
http://cell-nucleus-lab.bio.spbu.ru/people.html
Researchers from St Petersburg University and Friedrich-Schiller-Universität Jena have developed a unique technique of chromosome microdissection which allows scientists to obtain and decode even tiny DNA fragments. The research findings have been published in BMC Genomics, a top-rated open access, peer-reviewed journal that considers articles on all aspects of genome-scale analysis, functional genomics, and proteomics. Currently there is a wide range of cytogenetic and cytological approaches to studying chromosomes – mechanical microdissection among them. Using a fine glass needle mechanical microdissection technique isolates a single chromosome or their regions, which further enables amplification of isolated material and its subsequent molecular cytogenetic analysis or high-throughput sequencing. Chromosome microdissection has been widely used to identify chromosome, chromosomal rearrangements, both evolutionary and clinically significant, architecture of the cell nuclei and etc. Relatively small physical size of mitotic chromosomes, however, limited the use of the conventional chromosome microdissection for investigation of tiny chromosomal regions. Researchers from SPbU and Friedrich-Schiller-Universität Jena have broken ground on science by developing a workflow for microdissection of giant transcriptionally active lampbrush chromosomes followed by the preparation of whole-chromosome and locus-specific fluorescent in situ hybridization (FISH)-probes, high-throughput sequencing and mapping the smallest DNA fragments in genomes. "This new technique of chromosome microdissection isolates chromosomal regions as small in size as 1.5–4 mln base pairs", said Anna Zlotina, a lead author of the article, junior research associate at SPbU. "Now for successful amplification there is enough a tiny fragment of a single copy of chromosome, while previously in microdissection of metaphase chromosomes multiple copies, at least 10-15, of dissected regions, subsequently larger in size, were necessary". "The new workflow can be applied for mechanical microdissection of giant transcriptionally active lampbrush chromosomes", said Alla Krasikova, Associate Professor at SPbU. "The developed technical approach allows to obtain DNA and RNA samples from particular lampbrush chromosome loci, to define precisely the genomic position, extent and sequence content of the dissected regions. Technically unique in its kind and highly efficient, it allows scientists to correlate particular cytological structures and even individual genes with known DNA sequences. Also, it is convenient in performing in-depth analysis of functionally important chromosome regions and mechanisms of how the genetic information is realised". Read more at 'Microdissection of lampbrush chromosomes as an approach for generation of locus-specific FISH-probes and samples for high-throughput sequencing', BMC Genomics, 2016 (https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-016-2437-4).
The St. Petersburg team have also conducted a study in the topography of surfaces of nuclear structures. They have obtained unique surface images of the cell. It became possible due to the scanning electron microscope of the Centre for Nanotechnology at the SPbU Research Park. The results of the study have been published in Scientific Reports, a reputable international journal. SPbU biologists aimed to investigate ultrastructural topography of surfaces of nuclear structures – lampbrush chromosomes and nuclear bodies, in particular, nucleoli, histone locus bodies, Cajal bodies, and interchromatin granule clusters. To study intranuclear structures, the research team focused on nuclei of avian and amphibian oocytes, which frequently become models in biological studies of the cell nucleus. The large size of these nuclei allows to microsurgically isolate a range of intact nuclear bodies and lampbrush chromosomes into distinct preparations. Low-voltage scanning electron microscopy – a modification to the method of scanning electron microscopy – allowed to analyse uncoated, non-osmicated biological samples. According to Alla Krasikova, SPbU Associate Professor of the Department of Cytology and Histology, the research lies at the intersection of methods used in nuclear physic and cell biology. The analysis allowed to obtain unique high-resolution surface images of nuclear structures. "Apart from the morphology of nuclear domains, we examined the distribution of certain marker components. For that end, we used antibodies with subsequent detection of colloidal gold particles. The results of the study suggest that ultrastructural surface may reflect functional status of a nuclear body", says Alla Krasikova. Click on the link to learn more about the study 'Low-voltage scanning electron microscopy study of lampbrush chromosomes and nuclear bodies in avian and amphibian oocytes' (http://www.nature.com/articles/srep36878).
In the recent years, biologists have been able to identify the formation mechanisms of intranuclear structures. It turned out that the key role in their formation is played by RNA and RNA binding proteins. Most RNA binding proteins contain unstructured threads. The researchers at St. Petersburg hypothesise that the binding of such proteins and RNA may result in their aggregation (irreversible association) inside of the cell nucleus. At the same time, certain structural features of RNA binding proteins allow them to form dynamic drop-like formations: nuclear domains. In an article published in Chromosoma, St. Petersburg State University researchers provide evidence of the work of the mechanism of nuclear domain formation. The study analyses unusual nuclear domains – giant terminal RNP aggregates (GITERA). They are formed on certain avian lampbrush chromosomes and contain a specific set of RNA binding proteins, including components of the RNA processing mechanism. Thus, GITERA formed at the termini of pigeon lampbrush chromosomes contain proteins with unstructured threads: p54nrb and hnRNP I / PTB. RNA binding proteins containing unstructured threads or prion-like domains have the ability to aggregate and form amyloid-like fibres. It was previously proven that mutations in these regions of RNA binding proteins can lead to the formation of pathological aggregates in the cytoplasm and nuclei of human neurons and muscle cells in a number of neuromuscular degenerative diseases. It was assumed that the aggregate formation is an exclusively pathological feature. St. Petersburg State University researchers have reached the opposite conclusion: aggregate formation is possible in normal cells as well. The findings of SPbU scientists will make it possible to study this process in normal cells. This will help biologists to continue the study of the aggregation of proteins with unstructured threads; a process that can lead to neuro- and muscular degenerative diseases in humans. More on the results of the study: 'Giant poly (A)-rich RNP aggregates form at terminal regions of avian lampbrush chromosomes'. Chromosoma. 2015. 1–16 (http://link.springer.com/article/10.1007%2Fs00412-015-0563-4).
Our lab continues to study both the structure of LBCs and the RNA transcribed from them, using the favorable chromosomes of the newt Notophthalmus viridescens and the axolotl Ambystoma mexicanum for most of our structural studies, and the RNA from GVs of the frogs Xenopus tropicalis and X. laevis for our molecular analyses.
Over the past few years we have used super-resolution microscopy to examine individual transcription units of Notophthalmus in greater detail than is possible by confocal microscopy. Depending on the type of microscope used, we obtain resolutions in the range of 50-100 nm, compared to 200 nm for confocal. Although this resolution does not match what is possible by electron microscopy, the great advantage of the newer microscopes is that they are based on fluorescence. Thus, the full range of fluorescent tags and immunofluorescent antibodies is available.
Using antibodies against RNA polymerase II we see increased detail in the axes of individual LBC loops. Specifically, pol II is not uniformly distributed along the axis. Instead, there are short regions with polymerase staining interspersed with regions that lack polymerase. It is not clear, of course, how much of this pattern might be preparative artifact, but it is possible that there are regions where the polymerase moves rapidly along the axis and regions where it pauses.
We used the brilliant staining of an antibody against CELF1 protein to look at details of the nascent transcripts (see the 2007 Chromosome Research paper by Garry Morgan for why this protein is valuable for analyzing LBC loops). Super-resolution reveals that the coarsely granular pattern of CELF1 staining on certain loops is due to what appears to be rather uniform particles with a diameter no greater than about 100 nm. Again, the issue of artifacts must be considered, both from sample preparation and possibly from the algorithms used to generate the images. Our first publication using super-resolution microscopy appeared in Chromosome Research in 2012 (Kaufmann, Cremer, and Gall).
To complement our structural analysis of LBC loops, we embarked several years ago on a program to analyze the transcriptome of the oocyte by deep sequencing. We chose Xenopus tropicalis for this purpose, because its genome has been sequenced and annotated in greater detail than that of any other amphibian. A unique advantage of the Xenopus oocyte is that pure samples of cytoplasmic and nuclear RNA can be obtained by manual dissection, something that is almost impossible with any other cell type. We found that cytoplasmic RNA, as expected, contains spliced transcripts derived from thousands of genetic loci. Nuclear RNA, on the other hand, was a complete surprise. We expected to see nascent transcripts or partially processed pre-mRNA molecules (in addition to ribosomal RNA precursors and small RNAs of various sorts). Instead we found an abundance of stable RNAs derived from the introns of the same genes represented in the cytoplasm. We refer to this population as stable intronic sequence (sis) RNA (Gardner et al. 2012). Surprisingly, we find a population of sisRNA molecules in the cytoplasm of the oocyte, all of which are circular (lariats without tails) (Talhouarne and Gall 2014). We find that sisRNA is transmitted intact to the early embryo and persists through early embryogenesis until at least the blastula stage. So far we do not know the function(s) of sisRNA.
In retrospect, we should not have been surprised that we did not detect nascent transcripts in our GV RNA samples. Although transcription per gene is very high at the LBC stage, the GV contains only four copies of each gene (two in each homologue). Thus the total amount of RNA present as nascent transcripts in a single GV is vanishingly small, and our RNA samples are derived from only a few hundred hand-isolated GVs. By careful bioinformatic analysis, Zehra Nizami in our lab has begun to detect the minor fraction of nascent transcripts in our GV RNA samples. Eventually we hope to characterize these transcripts and to correlate our sequence data with what can be seen on the LBCs by in situ hybridization.

|
A remarkable career in science—Joseph G. Gall
|
The specimen was prepared by placing the contents of an oocyte nucleus in deionised water, thus causing dispersal of the nucleolar components; the unwound cores were centrifuged through a neutral solution of 0.1 molar sucrose with 10% formalin onto a carbon covered grid; the grid was rinsed in 0.4% Kodak Photo-Flo solution before drying; the preparation was then stained for 1 minute with 1% phosphotungstic acid in 50% ethanol at pH 2.5 (unadjusted), rinsed in 95% and then 100% ethanol, and dried with isopentane. (Miller, O.L. and Beatty B.R. 1969. Visualization of nucleolar genes. Science N.Y. 164, 955 – 957)
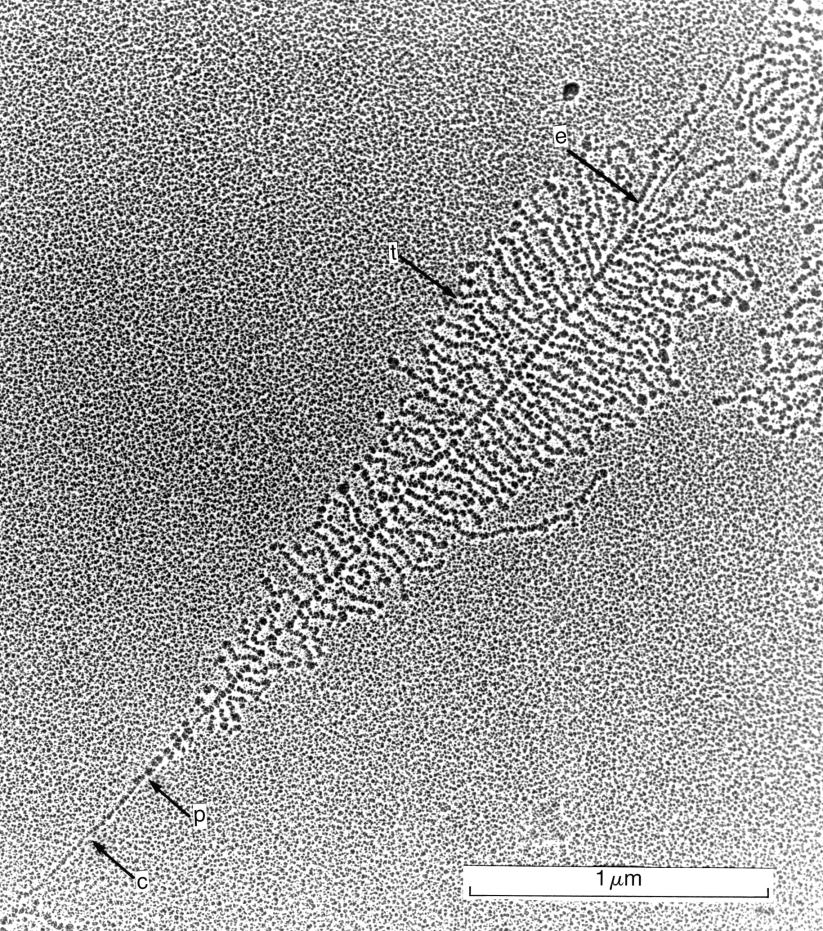
An electron micrograph of a single nucleolar gene transcription unit prepared by the Miller spreading technique. c: non-transcribing chromatin of the spacer region. p: the position of the first polymerase molecules at the start of the transcription unit. t: closely packed RNA transcripts extending sideways from the DNA axis. e: the end of the transcription unit.

Eric Davidson, Norman Chandler Professor of Cell Biology at the California Institute of Technology, died on September 1, 2015.
Dave McClay has written a rich and enormously worthy tribute to Eric to which I would not presume to add: http://www.sciencedirect.com/science/article/pii/S0012160615004005 (Developmental Biology, 406(2): 107-108, 15 October 2015).
It has to be said that the likes of Eric, myself and our contemporaries among the great scientists with whom we had the privilege to work were fortunate to enjoy what many of us regard(ed) as a golden age of scientific discovery: freedom to think, to experiment, to make bold assertions and have grand ideas and freedom to look at living systems outwith the constraints of "relevance" and "tenure" and "impact factor" and all the other impositions of modern research. Eric's contribution to biological science was massive and what's so special about it is that it came from a man who got a real kick out of his life and work.O One of my outstanding memories of him was at a Gordon Conference in the 1970s, when a group of us sat upon the ground and inadvertently consumed great quantities of American beer from dusk to dawn to the accompaniment of Eric on his banjo - the Golden Age indeed, of science!
As far as I know, he never made a preparation of lampbrush chromosomes. It's even possible that he never actually saw a live LBC. But he was the first person to draw attention to the need to know about all that RNA that was being made in the germinal vesicle. By studying the diversity and characteristics of the RNA in the eggs and germinal vesicles, he gave direction to our research and presented us with questions about gene expression and regulation that remain at the core of lampbrushology to this very day.
Eric was a great guy, loved and respected by all. He sets a fine example. If you're a young cell biologist, read about him and try to be a little bit like him.
|
|
|
|
|
|
News
|
 Webmaster
: Callum Macgregor,
UK
Webmaster
: Callum Macgregor,
UK
